When cancer SaaS solutions don’t work well in oncology data management, the problem is often integration. Genomic and lab data come in incorrect formats, like PDFs and LOINC codes that don’t match. Legacy EHR APIs make it hard to integrate into clinical workflows smoothly. This starts a chain reaction: oncologists lose timely insights, compliance teams are stuck under regulatory restrictions, AI software development teams struggle with missing data, and care coordination slows to a crawl.
To understand why these breakdowns persist, Jelvix analyzed more than fifty oncology-focused digital platforms. The work was motivated by a clear need: in an era of quantum leaps in AI and precision oncology, most platforms do not successfully operationalize innovation, as their integration layers are brittle.
By analyzing case studies, technical documentation, and workflow requirements (no personal, sensitive, or confidential data were used), this analysis revealed common challenges in data processing, EHR implementation, and compliance enforcement. The results demonstrated how state-of-the-art algorithms and clinical features can fail in the field when not supported by robust integration templates and resilient pipelines.
This research is based on publicly available technical documentation, product specifications, and platform materials from over 50 cancer-focused digital health platforms (no personal data or confidential information were used).
Why Integration Breaks Oncology SaaS Scale
From an analysis of over fifty oncology SaaS systems, we discovered two common ways in which these products fall short. The first is data heterogeneity: sequencing pipelines output structured files, pathology images are heavy in size, and clinical notes come in free text. Each requires a unique method for integration, yet most systems treat them as though they are the same streams.
The second is surface governance, with integrations that are stable only for pilots. The larger the platform (and/or the larger the number of platforms), the more sync errors and mismatched mappings, which makes it more difficult to onboard providers and damages the stability of AI modules in production.
These risks are made more serious by the bigger picture of the sector. Epic’s aggregation of cancer centers has made vendor lock-in hard to break, which makes EHR embedding a lot more complex. Meanwhile, changes in leadership and bankruptcy among oncology tech startups show a bitter truth: businesses that don’t plan for the expense of integration resiliency usually don’t make it. Even teams with great budgets fall apart when dysfunctional data pipelines begin to affect compliance, operations, and revenue.
The pattern is not new. From multimodal research pipelines powered by Graph Neural Networks to the rise and fall of Watson for Oncology, the evidence is consistent. No matter how advanced the algorithm, AI in oncology cannot compensate for broken integration. Oncology data management must be engineered from ingestion through analytics with resilience and real-time reliability in mind. Only then can SaaS platforms deliver the continuous, compliant, AI-ready infrastructure that modern oncology care demands.
Struggling to prepare healthcare data for your AI? Learn how to build a reliable data pipeline and accelerate your solutions with our proven guide.
The True Cost of Fragmented Data in Oncology Tech
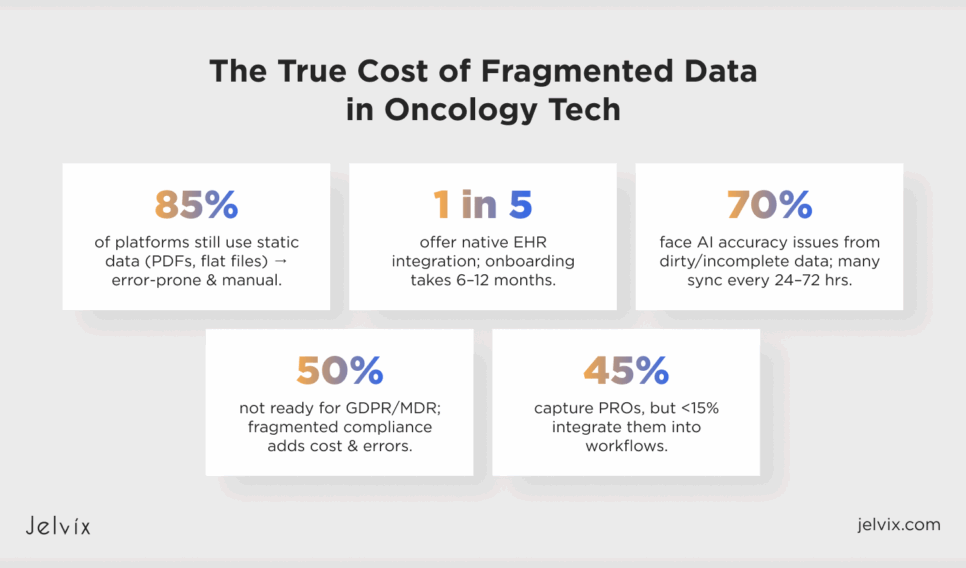
A review of more than fifty oncology SaaS platforms reveals that most solutions remain constrained by legacy data practices and fragile integration strategies. Most, almost 85 percent, continue to receive and manage genomic and pathology results as static data in formats that include scanned PDFs, flat files, or vendor portals. Yet, there are established structured standards (e.g., LOINC, HL7, and FHIR); some of them are used with variable adoption and almost always with manual curation or custom scripts that leave the door open to mistakes. The result is easy to forecast: instead of being designed with scale in mind, oncology software ends up relying on fragile pipelines that can’t support real-time workflows.
These vulnerabilities often become more pronounced in the context of EHR integration. Industry observations suggest that only a fraction of platforms, perhaps one in five, provide native embedding into Epic, Cerner, or similar systems.
Lots of them rely instead on exported summaries or external portals, which could slow down the oncologists, as they have to jump back and forth between those different tools. 58% of organizations believe EHR onboarding is a top bottleneck, and it takes six to 12 months to integrate, according to a survey. The absence of well-recognized endpoints for cancer-related data fields, which may include genomic variants or biomarker panels, might exacerbate delays, and even well-prepared deployments might suffer from prolonged stalls.
Upstream failures are plaguing artificial intelligence, which has been touted as a game-changer for oncology. Approximately seventy percent of the platforms reviewed indicated challenges with model accuracy, which appeared to stem from issues such as dirty data, incomplete pipelines, or limited traceability.
Models designed to predict recurrence or recommend biomarker-driven therapy fail to meet expectations, as the inputs are nonspecific. A number of participants are still on batch synching schedules of 24-72 hours, resulting in further delays from timeliness and leading to care-coordination delays that sabotage AI-generated recommendations.
The compliance dimension is equally troubling. More than half of the systems reviewed appear not to be out-of-the-box ready for multi-regional regulations such as GDPR or MDR, which can create costly re-architecture efforts when scaling beyond the U.S. In practice, standards like HIPAA, GDPR, and ONC are often implemented separately, which may result in duplicated administration and fragmented approaches to logging and auditing. This split also introduces more opportunities for error and bogs down product iteration.
Patient-reported outcomes (PROs) provide another example of unrealized potential. According to our analysis of public-facing documentation, around 45% of oncology SaaS solutions appear, in theory at least, to offer a PROs capture with symptom trackers, pain logs, or questionnaires. Yet, less than 15% appear to recently support integration into clinical workflows (like PROs injection into decision-making or real-time alerts).
Taken together, the findings indicate that the oncology SaaS ecosystem continues to struggle with core infrastructural issues rather than algorithmic sophistication. The patterns cut across genomics interpretation tools, tumor board software, clinical decision support platforms, oncology AI engines, trial-matching platforms, and cancer registries with SaaS dashboards. All face this fundamental truth: ingestion methods have been static, EHRs have been integrated late or not at all, AI underperforms in dirty data environments, compliance has been fragmented, and PRO data has not been operationalized.
However, the FHIR interoperability standard, cloud-native pipelines, and a patient-centric data model are all showing that the problem is solvable. Platforms that do not treat integration resilience as subterranean infrastructure but rather as core building blocks are already demonstrating accelerated provider onboarding and more robust AI deployments.
How To Design Resilient Integration in Oncology Medical Software
The journey to resilience in oncology integration begins with reimagining architecture. The core of all platforms depends on fragile point-to-point connections that simply break down as the sources of data scale. A more powerful approach is the cancer data mesh: a thin layer of distributed labs, genomics archives, imaging archives, and oncology EHR systems publish standardized data objects into a governed network. Rather than a single hub that gets bogged down under load, a mesh ensures a source owns a domain and a schema version.
For example:
- Genomic sequencing outputs into JSON bundles enriched with LOINC and SNOMED mappings;
- Imaging systems can push DICOM metadata with standardized identifiers into the same mesh.
This provides a single substrate for downstream analytics and AI pipelines, with no need for redundant ad hoc transformations.
The next layer of resilience is bi-directional EHR synchronization. Oncology medical software cannot rely on one-way HL7 feeds that update once daily. Instead, platforms should build event-driven FHIR APIs capable of handling continuous updates.
Integration resilience also depends on a unified architecture for labs, genomics, and AI pipelines. Oncology data arrives in mixed formats, like structured VCF files, semi-structured PDFs, and unstructured histopathology notes, and each requires specialized handling. A resilient ingestion pipeline should include:
- OCR and NLP engines to process pathology reports and clinical notes.
- Ontology-based mapping services to normalize identifiers across labs and clinical systems.
- Lineage and quality checks in a staging layer to guarantee clean downstream feeds.
- Distribution mechanisms that route validated data to oncology data analytics and AI pipelines.
This guarantees that transformer models working on text and graph neural networks consuming genomic-phenotypic associations both receive consistent, reliable inputs.
Ultimately, we must view interoperability in healthcare benefits, challenges, and resolutions through an engineering lens. The advantages are obvious: real-time coordination, quicker provider onboarding, and predictable AI deployment. The issues are just as apparent: outdated EHR APIs, disparate lab data, regulatory barriers, and incomplete audit trails.
The resolutions demand fine-grained cancer data meshes, subscription FHIR APIs, cloud-native staging layers, and schema registries that enforce structure at scale. Platforms that bake these principles into the architecture graduate from brittle integrations to robust, AI-enabled infrastructure.
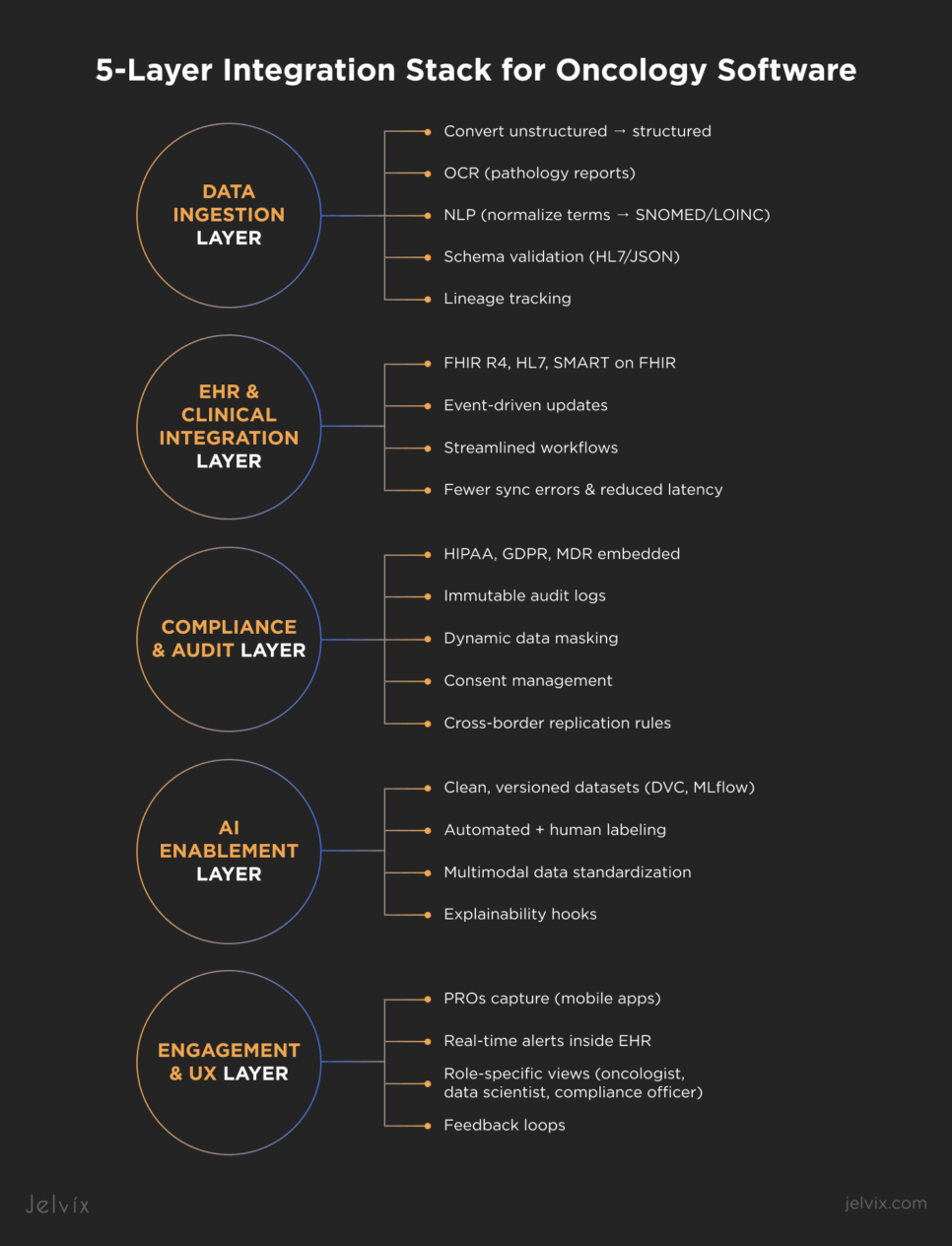
Building the 5-Layer Integration Stack for Oncology Software
Scaling oncology software requires a layered integration stack. By structuring integrations into five distinct layers, oncology SaaS platforms can move beyond brittle pilots and deliver resilient pipelines that empower everyone involved.
Data Ingestion Layer: Unstructured to Structured
The foundation of integration resilience begins at ingestion. Oncology data arrives in unstructured forms, including PDF pathology reports, free-text clinician notes, raw VCF genomic files, or imaging outputs in DICOM. The ingestion layer converts these into structured, normalized formats suitable for downstream use. An oncology data specialist typically employs:
- OCR engines to extract structured content from scanned pathology reports.
- NLP pipelines fine-tuned on oncology corpora to normalize terminology and map synonyms to SNOMED or LOINC.
- Schema validation services that reject malformed HL7 or JSON payloads before polluting production systems.
- Lineage tracking to guarantee that each genomic variant or lab value can be traced back to its source.
EHR and Clinical Integration Layer (FHIR, HL7, SMART)
The second layer aligns oncology software with clinical workflows. Relying solely on HL7 v2 interfaces isn’t sufficient. Resilient platforms must support FHIR R4 resources. They also support SMART on FHIR apps and event-driven subscriptions for continuous updates.
This eliminates duplicate data entry while also securing interoperability in healthcare. Such a layer addresses challenges and resolutions that providers demand: streamlined workflows, fewer sync errors, reduced latency, and improved trust in system outputs.
Compliance and Audit Layer
Without integration-level adherence, oncology software cannot scale between regions. A nimble compliance layer plugs HIPAA, GDPR, and MDR requirements directly into your data pipelines.
Core features include:
- Immutable audit logs of every data touchpoint.
- Dynamic data masking for concealing patient-specific information that is yet useful for analyses.
- Patient portals with consent management services to capture and enforce data sharing preferences.
- Cross-border replication rules that ensure data replication does not go to unauthorized jurisdictions.
By operationalizing compliance, oncology data specialists reduce manual reporting overhead and accelerate expansion into regulated markets.
AI Enablement Layer (Clean Labeling, Versioning)
AI in oncology is only as reliable as the pipelines feeding it. The AI enablement layer transforms data into training- and inference-ready datasets.
This requires:
- Data versioning system: DVC or MLflow to manage evolving datasets.
- Automated labeling pipelines combining rule-based classifiers and human-in-the-loop validation for ambiguous cases.
- Model input standardization, where imaging, clinical, and genomic oncology data are converted into interoperable formats for multimodal learning.
- Explainability hooks so data analytics outputs can be traced back to source inputs for regulatory alignment.
Without this layer, AI modules overfit to noise and fail in production.
Engagement & UX Layer (PROs, Alerts)
The top layer ensures that integrated data flows back into experiences that oncologists and patients actually use. Modern oncology software cannot stop at analytics dashboards. It must embed integration outputs into daily workflows.
This includes:
- Patient-reported outcomes (PROs) collected via mobile apps and fed into the clinical data stream.
- Real-time alerts delivered inside the EHR context.
- Role-specific views where oncologists see treatment pathways, data scientists access analytics-ready streams, and compliance officers monitor audit trails.
- Feedback loops that capture clinician interaction patterns and refine pipeline performance over time.
By layering engagement on top of ingestion, integration, compliance, and AI readiness, oncology software delivers measurable value to every stakeholder.
Tired of fragmented oncology data? See how our Data Management Solution helped a SaaS leader consolidate data for real-time analytics.
The Role of AI in Oncology Data Management—and Why It Fails
While promising, artificial intelligence has yet to show reliable clinical utility in oncology. Many deployments seem to struggle less with the underlying algorithms and more with the fragile data infrastructures that support them. When models are trained on semi-structured PDFs, partial EHR extracts, or poorly formatted lab values, the result is predictable: noisy inputs yield unreliable outputs. For oncologists, these failures manifest as hallucinated predictions or incomplete recommendations, eroding clinical trust and undermining adoption.
Resilient AI in oncology requires the same engineering rigor that drives enterprise-scale healthcare software development. That discipline begins with traceability. A model must be able to trace back every clinical note, genomic variant, and pathology image it uses to its original source, together with information about quality, consent, and the time it was taken. Without lineage, it is impossible to explain things, and regulators have no way to check recommendations.
The second requirement is clean interoperability. Genomic pipelines quintessentially need to output structured VCF variants that are aligned with standardized ontologies. DICOM metadata with stable identifiers is required by imaging systems. An FHIR integration solution will be required for clinical data to be passed through, which supports real-time subscriptions. In such an environment, when an oncologist modifies a treatment order, the change is propagated instantly downstream, enabling AI models to adjust inference without retraining on outdated datasets.
And lastly, resilience is the result of a closed-loop feedback. There can be no vacuum of AI software development in oncology; models need to capture real-world outcomes and clinician overrides and feed them back into the training set. This circuit prevents fixation and promotes improvement in the quality of prediction over time. Complementary to strong FHIR pipelines, it makes for a resilient system that keeps the AI outputs synchronized with both the clinical world and regulatory landscape.
The lesson is simple but frequently missed: failures of AI in oncology stem less from inadequacies in modeling than from weaknesses in the supporting infrastructure. Applied with disciplined engineering—lineage tracing, real-time interoperability, and constant feedback—the effects are dramatic: AI stops hallucinating and starts subjecting itself to lipid tests, delivering the precision insights oncologists hope for.
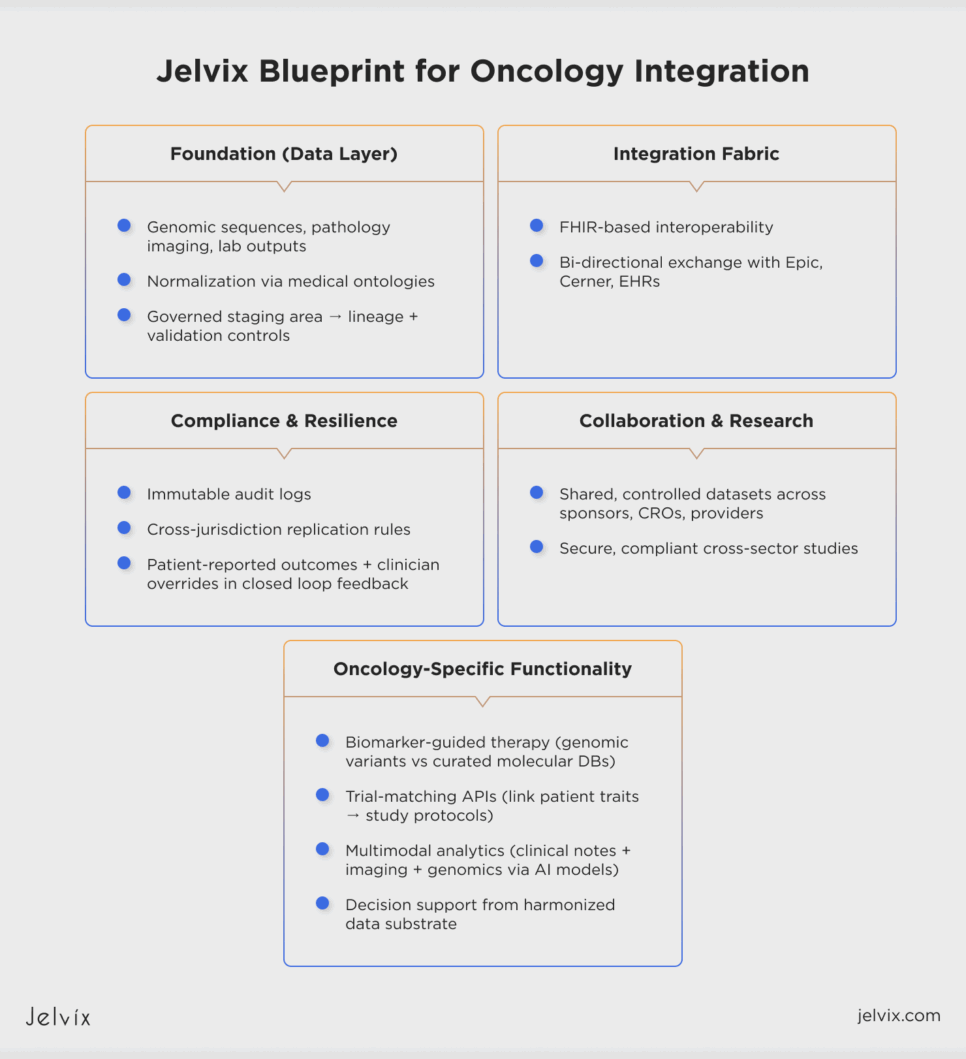
Jelvix Blueprint for Oncology Integration
Jelvix conceptualizes oncology integration as an architectural discipline in which durability must be embedded within the data layer rather than appended at the application tier. In this pipeline, genomic sequences, pathology imaging, and laboratory outputs are piped through normalization processes that are grounded in established medical ontologies.
They fuel a governed staging area under strict lineage and validation controls to deliver data integrity and provenance throughout the end-to-end use lifecycle. On this foundation, an FHIR-based integration fabric establishes interoperability with electronic health record ecosystems such as Epic and Cerner, enabling consistent bi-directional exchange of oncology-specific data.
The architectural design supports functionality uniquely required in oncology. Normalized genomic variants may then be compared systematically to existing curated molecular databases to aid biomarker-guided therapy decisions. Structured trial datasets can similarly be exposed via eligibility-oriented APIS to facilitate the linking of patient-specific genomic traits and a clinical research study protocol.
Additionally, multimodal data streams—unstructured clinical notes and image-derived metadata—are analyzed through state-of-the-art computational techniques (eg, transformer-based models or graph neural networks). In this setup, complex inferencing and decision support are realized via heterogeneous but harmonized data modalities through the platform. This lets oncology analytics work on a single substrate instead of separate silos.
Integration resilience is built in as a design requirement. Immutable audit records and cross-jurisdictional replication rules work under the analytics layer to make sure that regulatory obligations are always and automatically followed. Patient-reported outcomes and clinician overrides go back into the data stream through controlled channels, making a closed loop that improves both the accuracy of predictions and the integrity of compliance.
The architecture facilitates cross-sector studies where sponsors, CROs, and providers work together on shared but controlled datasets without putting security or compliance at risk by applying these similar concepts to clinical research data management services.
Checklist: Is Your Oncology SaaS Integration-Ready?
Answer each question with “Yes” or “No”:
Are genomic and lab results consistently normalized to standards such as LOINC and SNOMED?
Do ingestion pipelines enforce schema validation and lineage tracking with automated quality checks?
Have you implemented a FHIR integration solution with real-time subscriptions and bi-directional sync?
Are insights embedded directly inside EHR workflows and updated without duplicate entry?
Is compliance automated with audit logs, data masking, encryption, and jurisdictional controls?
Are datasets versioned and labeled in a standardized way that supports oncology analytics and AI pipelines?
Do treatment updates and lab results propagate across systems in minutes rather than days?
Are patient-reported outcomes and clinician overrides automatically captured and fed back into pipelines?
Does your architecture support clinical research data management services with secure and controlled sharing?
Has your integration layer been stress-tested for regional deployments, concurrent users, peak API calls, and failover recovery?
Scoring Your Readiness
9–10 “Yes” answers: Your platform is integration-ready and built for scale.
6–8 “Yes” answers: Your platform is functional but needs targeted improvements before scaling further.
3–5 “Yes” answers: Your integration stack is fragile; significant architectural adjustments are required.
0–2 “Yes” answers: Your platform is not integration-ready; foundational redesign is necessary.
If your oncology SaaS has a high score, it already has the strength to grow, but it may require some work to keep up with future needs. If you’re just partially ready, you could run into problems with onboarding providers, making sure you’re following the regulations, and making certain AI is reliable. Also, if your platform isn’t entirely ready, the issues are structural and need to be fixed in a planned way.
We know how to improve interoperability in healthcare at Jelvix. We can help you make your systems more interoperable and grow safely if you’re ready to integrate. We can also help you come up with the strategy and architecture you need to get there if you’re not ready for integration yet. Book your 30-minute call with the Jelvix experts team to discuss the integration foundation that your oncology platform needs.
Looking to scale your product with focus and speed?
Get a team of specialists committed solely to your success from kickoff through launch.